BRKGA: Biased Random Key Genetic Algorithm#
An excellent and very informative presentation about BRKGAs can be found here. BRKGAs are known to perform well on combinatorial problems.
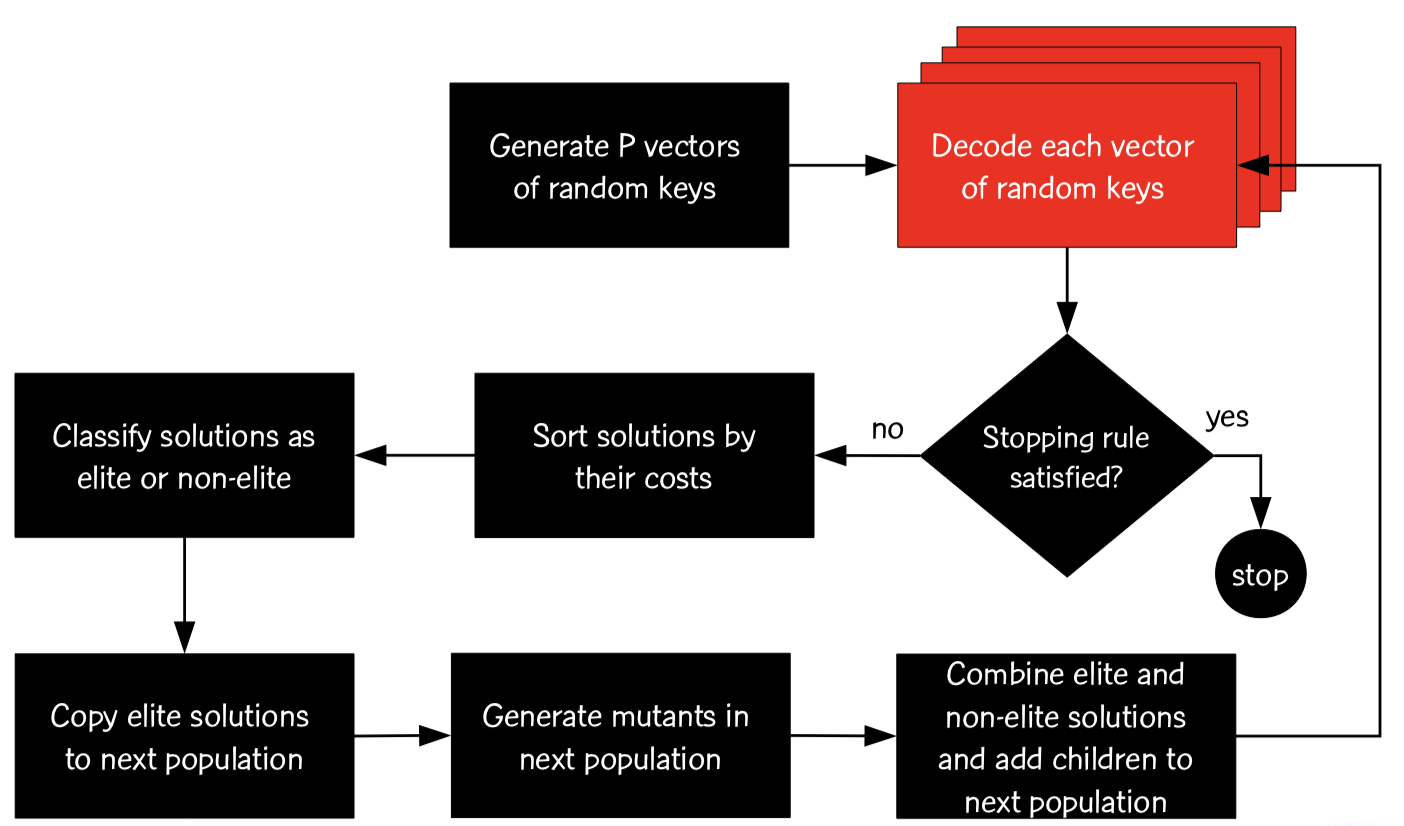
Instead of customizing evolutionary operators, decoding has to be defined. Therefore, evolution takes place solely on real-valued variables.
Let us define a permutation problem which derives an order by sorting real-valued variables:
[1]:
import numpy as np
from pymoo.core.problem import ElementwiseProblem
class MyProblem(ElementwiseProblem):
def __init__(self, my_list):
self.correct = np.argsort(my_list)
super().__init__(n_var=len(my_list), n_obj=1, n_ieq_constr=0, xl=0, xu=1)
def _evaluate(self, x, out, *args, **kwargs):
pheno = np.argsort(x)
out["F"] = - float((self.correct == pheno).sum())
out["pheno"] = pheno
out["hash"] = hash(str(pheno))
Since duplicate eliminates is an essential aspect for evolutionary algorithms, we have to make sure all duplicates with respect to the permutation (and not to the real values) are filtered out.
[2]:
from pymoo.core.duplicate import ElementwiseDuplicateElimination
class MyElementwiseDuplicateElimination(ElementwiseDuplicateElimination):
def is_equal(self, a, b):
return a.get("hash") == b.get("hash")
Then, we define a problem that has to sort a list by their values.
[3]:
np.random.seed(2)
list_to_sort = np.random.random(20)
problem = MyProblem(list_to_sort)
print("Sorted by", np.argsort(list_to_sort))
Sorted by [ 1 19 12 14 6 9 8 5 4 3 0 17 13 11 2 7 10 15 18 16]
Finally, we use BRKGA to obtain the sorted list:
[4]:
from pymoo.algorithms.soo.nonconvex.brkga import BRKGA
from pymoo.optimize import minimize
algorithm = BRKGA(
n_elites=200,
n_offsprings=700,
n_mutants=100,
bias=0.7,
eliminate_duplicates=MyElementwiseDuplicateElimination())
res = minimize(problem,
algorithm,
("n_gen", 75),
seed=1,
verbose=False)
print("Best solution found: \nX = %s\nF = %s" % (res.X, res.F))
print("Solution", res.opt.get("pheno")[0])
print("Optimum ", np.argsort(list_to_sort))
Best solution found:
X = [0.46254633 0.04439827 0.76134297 0.45693248 0.44636578 0.43021774
0.09891258 0.78862076 0.38532637 0.20650484 0.80437363 0.69087889
0.0767443 0.65504853 0.08779362 0.91798022 0.98682869 0.56451515
0.95299978 0.07605258]
F = [-20.]
Solution [ 1. 19. 12. 14. 6. 9. 8. 5. 4. 3. 0. 17. 13. 11. 2. 7. 10. 15.
18. 16.]
Optimum [ 1 19 12 14 6 9 8 5 4 3 0 17 13 11 2 7 10 15 18 16]
API#
- class pymoo.algorithms.soo.nonconvex.brkga.BRKGA(self, n_elites=200, n_offsprings=700, n_mutants=100, bias=0.7, sampling=FloatRandomSampling(), survival=None, output=SingleObjectiveOutput(), eliminate_duplicates=False, **kwargs)[source]
- Parameters:
- n_elitesint
Number of elite individuals
- n_offspringsint
Number of offsprings to be generated through mating of an elite and a non-elite individual
- n_mutantsint
Number of mutations to be introduced each generation
- biasfloat
Bias of an offspring inheriting the allele of its elite parent
- eliminate_duplicatesbool or class
The duplicate elimination is more important if a decoding is used. The duplicate check has to be performed on the decoded variable and not on the real values. Therefore, we recommend passing a DuplicateElimination object. If eliminate_duplicates is simply set to True, then duplicates are filtered out whenever the objective values are equal.