Parallel Coordinate Plots#
For higher-dimensional data, Parallel Coordinate Plots are a powerful technique to analyze how dense solutions are distributed in different ranges regarding each coordinate.
Let us create some data for visualization:
[1]:
from pymoo.problems.many.dtlz import DTLZ1
from pymoo.util.reference_direction import UniformReferenceDirectionFactory
ref_dirs = UniformReferenceDirectionFactory(6, n_partitions=5)() * [2, 4, 8, 16, 32, 64]
F = DTLZ1().pareto_front(ref_dirs)
This is the Pareto-front for the DTLZ1 test problem with six objectives, with some scale added. We add a different scaling to show the effect of normalization later on. Let us assume our algorithm converged after some generations, and this is the result set.
[2]:
from pymoo.visualization.pcp import PCP
PCP().add(F).show()
[2]:
<pymoo.visualization.pcp.PCP at 0x1090a5790>
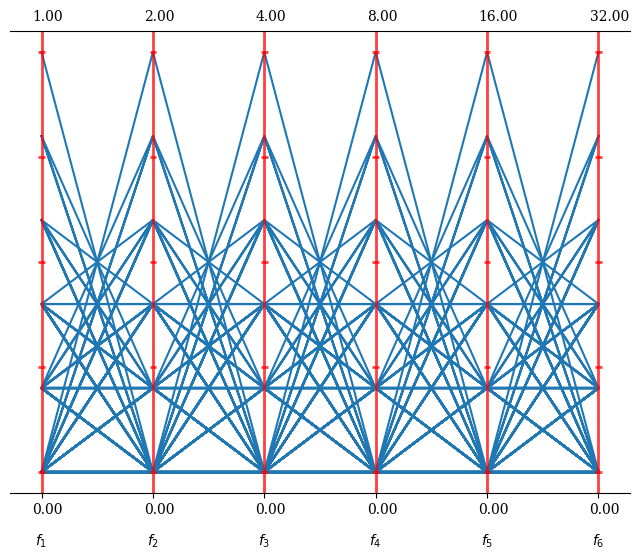
This gives an idea of the overall result set. Let us assume we identified solution 50 and 75 to be more of interest and would like to highlight them in our plot:
[3]:
plot = PCP()
plot.set_axis_style(color="grey", alpha=0.5)
plot.add(F, color="grey", alpha=0.3)
plot.add(F[50], linewidth=5, color="red")
plot.add(F[75], linewidth=5, color="blue")
plot.show()
[3]:
<pymoo.visualization.pcp.PCP at 0x1096377d0>
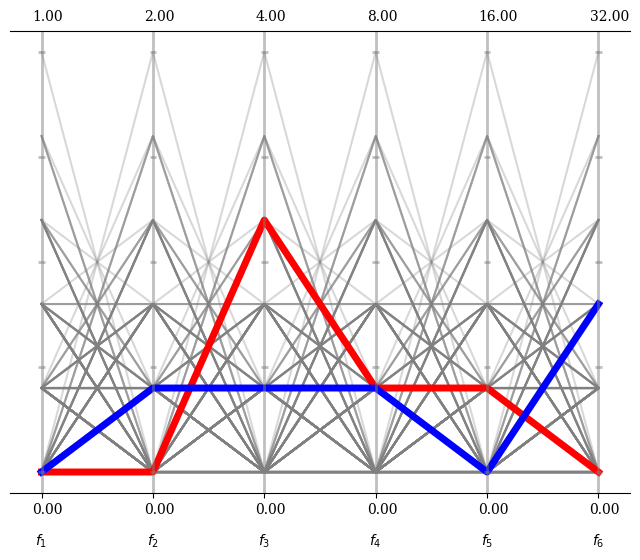
Please note that the PCP object just is a wrapper around a matplotlib figure. All options that apply for plotting the corresponding type (here plot, but it can also be scatter, polygon, …) can be used, such as linewidth, color or alpha.
Some more options to be used in a plot
[4]:
plot = PCP(title=("Run", {'pad': 30}),
n_ticks=10,
legend=(True, {'loc': "upper left"}),
labels=["profit", "cost", "sustainability", "environment", "satisfaction", "time"]
)
plot.set_axis_style(color="grey", alpha=1)
plot.add(F, color="grey", alpha=0.3)
plot.add(F[50], linewidth=5, color="red", label="Solution A")
plot.add(F[75], linewidth=5, color="blue", label="Solution B")
plot.show()
[4]:
<pymoo.visualization.pcp.PCP at 0x109711bb0>
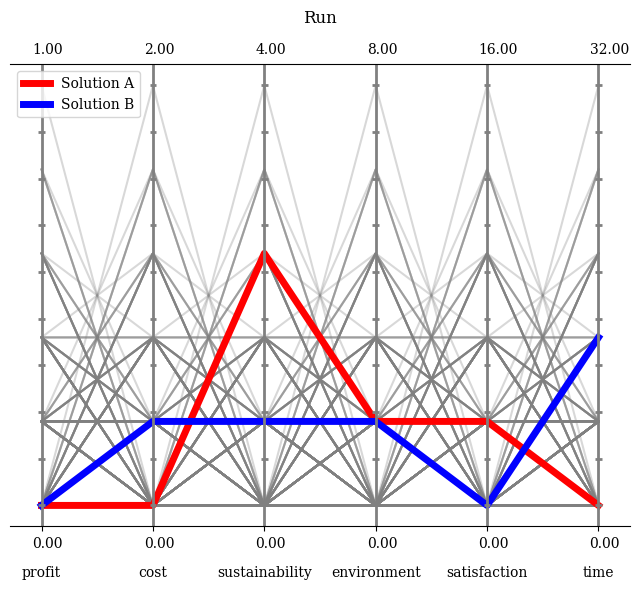
Moreover, if the boundaries should be set manually, this can be achieved by turning the default normalization off and providing them. Either directly as a NumPy array or just an integer to be set for all axes.
[5]:
plot.reset()
plot.normalize_each_axis = False
plot.bounds = [[1,1,1,2,2,5],[32,32,32,32,32,32]]
plot.show()
[5]:
<pymoo.visualization.pcp.PCP at 0x109711bb0>
<Figure size 800x600 with 0 Axes>
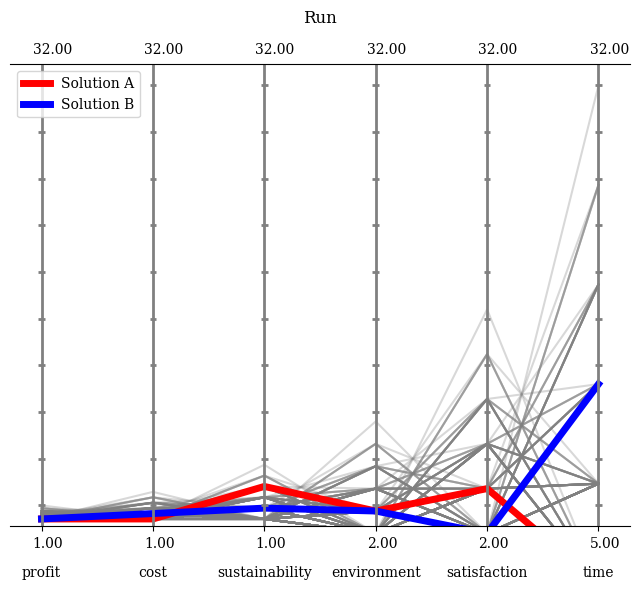
API#
- class pymoo.visualization.pcp.PCP(self, bounds=None, show_bounds=True, n_ticks=5, normalize_each_axis=True, bbox=False, **kwargs)[source]
Parallel Coordinate Plot
- Parameters:
- boundstuple
If plot requires normalization, it might be necessary to supply the boundaries. (Otherwise they might be approximate by the minimum and maximum of the provided data). The boundaries should be provided as a list/tuple or 2D numpy array, where the first element represents the minimum, second the second the maximum values. If only an integer or float is supplied, the boundaries apply for each variable.
- axis_styledict
Most of the plots consists of an axis. The style of the axis, e.g. color, alpha, …, can be changed to further modify the plot appealing.
- labelsstr or list
The labels to be used for each variable provided in the plot. If a string is used, then they will be enumerated. Otherwise, a list equal to the number of variables can be provided directly.
- n_ticksint
Number of ticks to be shown on each parallel axis.
- show_boundsbool
Whether the value of the boundaries are shown in the plot or not.
- normalize_each_axisbool
Whether the values should be normalized either by bounds or implicitly.
- Other Parameters:
- figsizetuple
The figure size. Default (figsize=(8, 6)). For some plots changing the size might have side effects for position.
- titlestr or tuple
The title of the figure. If some additional kwargs should be provided this can be achieved by providing a tuple (“name”, {“key” : val}).
- legendstr
Whether a legend should be shown or not.
- tight_layoutbool
Whether tight layout should be used.
- cmapcolormap
For some plots different kind of colors are used. The colormap can be changed to modify the color sequence for the plots.